DataFrames are essentially multidimensional arrays with attached row and column labels and sometimes infamous missing data.
Data Indexing and Selection
Series as dictionary, the Series object provides a mapping from a collections of keys to a collections of vlaues.
data = pd.Series([0.25, 0.5, 0.75, 1.0],
index=['a', 'b', 'c', 'd'])
data['b']
data.keys()
list(data.items())
#add a new index value
data['e']=1.25
We can also slice the data by the index like data[1:3] Or we can use the locattribute, data.loc[‘a’], the result would be 0.25. loc and iloc are very useful for cleaning data. Imagine we can a small dataframe using Pd.Series.
area = pd.Series({'California': 423967, 'Texas': 695662,
'New York': 141297, 'Florida': 170312,
'Illinois': 149995})
pop = pd.Series({'California': 38332521, 'Texas': 26448193,
'New York': 19651127, 'Florida': 19552860,
'Illinois': 12882135})
data = pd.DataFrame({'area':area, 'pop':pop})
data
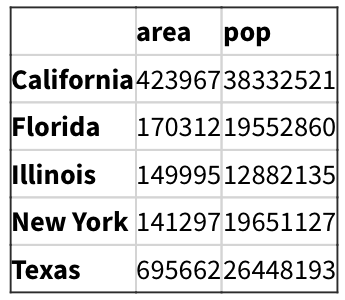
#Add another column
data['density'] = data['pop']/data['area']
#OMG, isnt it easy, its just like deal with csv file
We can also use data.value to swith this dataframe to a two-dimentional array([][]).
Little quizz, How do u pick density bigger than 100 and u only need column pop and density?data.loc[data.density > 100, ['pop', 'density']]
Handling Missing value
Datasets in real always have some amount of data missing and different data sources may indicate missing data in different ways. In this section, we will discuss some general considerations for missing data and how pandas chooses to represnet it. We will refer missing data in general as null,NaN,NAvalues.
Generally, there are two strategies: using a mask that globally indicate missing vlaues or choosing a sentinel value that indicates a missing entry.
In our case, we would use sentinel values cuz the first method may has some trouble(the overhead in both storage, computation and code maintenance).
The first sentinel value in pyhon is None, only used in arrays with data type object.
vals1 = np.array([1, None, 3, 4])
But the use of Python objects in an array also means when we perform aggreagations like sum() or min(), we would get error.
The next one is NaN(not a number),Keep in mind that NaN is specifically a floating-point value; there is no equivalent NaN value for integers, strings, or other types.
Whenever you use sum() or min(), the result would still be NaN.
Lets check on those missing values.
isnull(): Generate a boolean mask indicating missing values
dropna(): Return a filtered version of the data
data.isnull()
data.dropna() #drop any rows with missing values, so harsh
Time to fill those bitches!!! Sometimes rather than dropping NA values, you’d rather replace them with a valid value. The value might be a single number like zero or might be some sort of imputation or interpolation from the good values. We can use data.fillna(0) or we can We can specify a forward-fill to propagate the previous value forward data.fillna(method='ffill')
Hierarchical Indexing
We can use fuction MultiIndex to deal with tuples
index = [('California', 2000), ('California', 2010),
('New York', 2000), ('New York', 2010),
('Texas', 2000), ('Texas', 2010)]
populations = [33871648, 37253956,
18976457, 19378102,
20851820, 25145561]
pop = pd.Series(populations, index=index)
We can the index is a tuple, normally, we would do this to select value, pop[[i for i in pop.index if i[1] == 2010]]. That is super inefficient. We can do in this way instead.
index = pd.MultiIndex.from_tuples(index)
pop = pop.reindex(index)
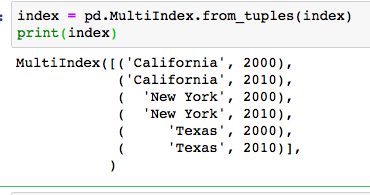 Then the tuble index would become two columns index. After that, we can just make a extre column out of it by
Then the tuble index would become two columns index. After that, we can just make a extre column out of it by pop.unstack()
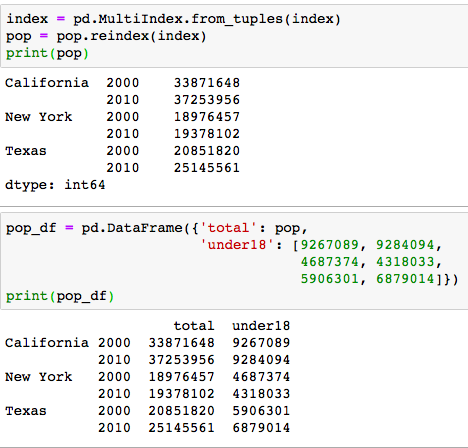 We can see from the photo, it is super nice to add another column with the index,eh?
We can see from the photo, it is super nice to add another column with the index,eh?
Combining Dataset